Codes of Chemical Structures: SMILES, InChI, and SMARTS
- Emre Can Buluz

- May 31, 2025
- 5 min read
Updated: Jun 10, 2025
The digital representation of chemical compounds is one of the cornerstones of modern chemistry, bioinformatics, and drug discovery studies. To serve this purpose, systems such as SMILES (Simplified Molecular Input Line Entry System), InChI (International Chemical Identifier), and other molecular identifiers have been developed, allowing structural information of molecules to be expressed in text format. These systems enable compounds to be searchable, comparable, and analyzable through software tools in databases.
SMILES Representation
In the SMILES (1) notation, atoms are represented by their atomic symbols. Aliphatic atoms are written in uppercase letters, while aromatic atoms are written in lowercase. SMILES is a hydrogen-suppressed representation, meaning that hydrogen atoms are usually not explicitly shown. Double bonds are represented by “=”, triple bonds by “#”, and single and aromatic bonds are generally not indicated with any symbol—except in certain cases like non-aromatic single bonds in biphenyl, where a “−” symbol may be used.
To generate a SMILES string, one must "walk" through the chemical structure in a way that each atom is visited only once. The simplest SMILES is likely for methane: C, where all four bonded hydrogens are implied. Ethane is written as CC, propane as CCC, and 2-methylpropane as CC(C)C (note the branching point). Cyclohexane demonstrates the use of ring closure numbers and is represented as C1CCCCC1. Benzene, with aromatic atoms, is written as c1ccccc1 (note the use of lowercase letters for aromatic atoms). Acetic acid is represented as CC(=O)O (2).
Information about chirality and geometric isomerism can also be specified in SMILES notation. Stereochemistry at chiral atoms is indicated using the “@” symbol. For example, the two stereoisomers of alanine are written as NC@HC(=O)O and NC@@HC(=O)O. Note that the hydrogen atom on the chiral carbon is explicitly included to define the stereocenter. Geometric isomerism (E/Z or cis-trans) around double bonds is represented using slashes. For example, trans-butene is written as C/C=C/C, while cis-butene is written as C/C=C\C.

InChI Representation
The goal of the IUPAC Chemical Identifier Project (IChIP) was to develop the IUPAC International Chemical Identifier (InChI), a unique label for chemical substances. This identifier serves as a non-proprietary tag for chemicals and can be used in both printed and electronic data sources. In this way, it facilitates easier linking across different datasets and ensures the clear and unambiguous identification of chemical substances.
InChI uses a layered format to represent all available structural information related to a compound’s identity. Each layer in the InChI representation contains specific structural data. These layers are generated from the input structure by extracting data automatically, and each successive layer is designed to add more detail to the identifier. The exact layers generated depend on the level of structural detail available and whether tautomerism is permitted. The sequential layers of an InChI are characterized as follows (3):
Formula
Connectivity (no formal bond orders)
a. Disconnected metals
b. Connected metals
Isotopes
Stereochemistry
a. Double bond
b. Tetrahedral
Tautomers (open or fixed)
An example of an InChI representation is given in Figure 2. It is important to note that InChI strings are designed for use by computers, and end-users are not expected to understand their details. InChI should be thought of like barcodes. In fact, the open structure and flexible representation of InChI allow software systems to integrate them in ways that enable scientists to worry less about the details of structure representation handled by computers.
The layers in an InChI string are separated by a slash (/) followed by a lowercase letter (except for the first layer, which is the chemical formula). The layers are arranged in a predefined order (3).
For guanine, an InChI version number is followed by the layers:
/ chemical formula
/c connectivity-1.1 (excluding terminal hydrogens)
/h connectivity-1.2 (locations of terminal hydrogens, mobile H addition points)
/q charge
/p proton balance
/t tetrahedral parity
/m parity inversion for relative stereochemistry
(1 = inverted, 0 = not inverted)
/s stereochemical type (1 = absolute, 2 = relative, 3 = racemic)
/f chemical formula of fixed-H structure, if different
/h connectivity-2 (locations of fixed mobile Hs)

One of the most important applications of InChI is that a chemical substance can be found using internet-based search engines. This is further facilitated by the use of the InChIKey. The InChIKey is a 27-character representation that is compressed and therefore cannot be converted back to the original structure, but it is protected against the unwanted and unpredictable truncation of longer character strings by some search engines.The usefulness of the InChIKey as a search tool is further increased if it is derived from a ‘standard’ InChI, that is, generated with standard option settings for properties like tautomerism and stereochemistry (3).
SMARTS Representation
The SMARTS language, invented by Daylight Information Systems in the late 1980s, has become an almost standard language for the definition of chemical representations. Although not complete, SMARTS is highly featured and allows chemists to precisely define a structural pattern that they have in mind (4). In the SMILES language, there are two basic symbol types: atoms and bonds. Using these SMILES symbols, a molecule’s graph (nodes and edges) can be defined, and labels can be assigned to the graph’s components (that is, specifying which atom type each node represents and which bond type each edge represents).The situation is similar in SMARTS: A graph is defined using atomic and bond symbols. However, in SMARTS, these node (atom) and edge (bond) labels are extended with logical operators and special atomic/bond symbols; this allows SMARTS atoms and bonds to be defined in more general ways. For example, [C,N] represents an aliphatic carbon or an aliphatic nitrogen atom; the tilde ~ symbol matches any bond type (5).
In SMARTS representation, atoms are indicated with square brackets ([]). For example:
[C]: Carbon atom
[N+]: Positively charged nitrogen
[O-]: Negatively charged oxygen
[c]: Aromatic carbon
Bonds are represented as follows:
- single bond
= double bond
# triple bond
: aromatic bond
~ any bond
@ chiral center
Examples of SMARTS:
[OH]: Hydroxyl group
[NX3;H2]: Primary amine (trivalent nitrogen with two H attached)
[#6]~[#8]: Any bond between carbon and oxygen
[c][nH]: Aromatic nitrogen bonded to aromatic carbon
[C;!R]: Carbon that is not in a ring

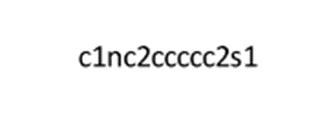
Molecular identifiers are among the most fundamental building blocks of the chemical information age. Each system—such as the practicality of SMILES, the standardized structure of InChI, and the flexible search power of SMARTS—enables molecules to be effectively represented and analyzed in the digital world. Thanks to these identifiers, chemical data becomes more accessible, comparable, and reproducible. With the advancement of software and data science technologies, the importance of these systems is growing every day; they play central roles in fields such as computational chemistry, drug design, and AI-assisted molecule discovery. Therefore, understanding the logic and applications of these identifiers has become an indispensable skill for anyone working with modern chemistry.
References
1. Weininger D, A Weininger and J L Weininger (1989). SMILES. 2. Algorithm for Generation of Unique SMILES Notation. Journal of Chemical Information and Computer Sciences 29:97–101.
2. Andrew R. Leach, V.J. Gillet. (2003). An Introduction to Chemoinformatics. Springer Science & Business Media.
3. Heller, S., McNaught, A., Stein, S., Tchekhovskoi, D., & Pletnev, I. (2013). InChI - the worldwide chemical structure identifier standard. Journal of cheminformatics, 5(1), 7. https://doi.org/10.1186/1758-2946-5-7
4. Ehrt, C., Krause, B., Schmidt, R., Ehmki, E. S. R., & Rarey, M. (2020). SMARTS.plus - A Toolbox for Chemical Pattern Design. Molecular informatics, 39(12), e2000216. https://doi.org/10.1002/minf.202000216
5. James C A, D Weininger and J Delany (2002). Daylight Theory Manual. Also at http://www.daylight.com/dayhtml/doc/theory/theory.toc.html.
6. Mellor, C. L., Steinmetz, F. P., & Cronin, M. T. (2016). Using Molecular Initiating Events to Develop a Structural Alert Based Screening Workflow for Nuclear Receptor Ligands Associated with Hepatic Steatosis. Chemical research in toxicology, 29(2), 203–212. https://doi.org/10.1021/acs.chemrestox.5b00480



Comments